Probability Sensitivity Analysis (PSA)
Nathan Green
2022-08-17
Source:vignettes/probability-sensitivity-analysis.Rmd
probability-sensitivity-analysis.RmdIntroduction
PSA is a core part of any cost-effectiveness analysis (Briggs et al. 2012). Here we will carry this out for a simple decision tree. This involves repeatedly sampling from a distribution for each branch probability and cost and calculating the total expected value for each set of realisations.
Example
library(CEdecisiontree, quietly = TRUE)
library(assertthat, quietly = TRUE)
library(treeSimR, quietly = TRUE)
library(tibble, quietly = TRUE)
library(tidyverse, quietly = TRUE)
library(purrr, quietly = TRUE)We first define the decision tree. The difference to previous trees is that we now use the list-column feature to define distributions rather than point values.
tree_dat <-
list(child = list("1" = c(2, 3),
"2" = NULL,
"3" = NULL),
dat = tibble(
node = 1:3,
prob =
list(
NA_real_,
list(distn = "unif", params = c(min = 0, max = 1)),
list(distn = "unif", params = c(min = 0, max = 1))),
vals =
list(
0L,
list(distn = "unif", params = c(min = 0, max = 1)),
list(distn = "unif", params = c(min = 0, max = 1)))))
tree_dat
#> $child
#> $child$`1`
#> [1] 2 3
#>
#> $child$`2`
#> NULL
#>
#> $child$`3`
#> NULL
#>
#>
#> $dat
#> # A tibble: 3 × 3
#> node prob vals
#> <int> <list> <list>
#> 1 1 <dbl [1]> <int [1]>
#> 2 2 <named list [2]> <named list [2]>
#> 3 3 <named list [2]> <named list [2]>We can now loop over this tree and generate samples of values for the
given distributions. We use the sample_distributions()
function from my treeSimR package. This is all wrapped up
in the convenience function create_psa_inputs.
library(purrr)
tree_dat_sa <- create_psa_inputs(tree_dat, n = 1000)This results in a list of trees.
head(tree_dat_sa, 2)
#> [[1]]
#> $child
#> $child$`1`
#> [1] 2 3
#>
#> $child$`2`
#> NULL
#>
#> $child$`3`
#> NULL
#>
#>
#> $dat
#> node prob vals
#> 1 1 NA 0.0000000
#> 2 2 0.9626497 0.2399404
#> 3 3 0.5228080 0.3990208
#>
#> attr(,"class")
#> [1] "tree_dat" "list"
#>
#> [[2]]
#> $child
#> $child$`1`
#> [1] 2 3
#>
#> $child$`2`
#> NULL
#>
#> $child$`3`
#> NULL
#>
#>
#> $dat
#> node prob vals
#> 1 1 NA 0.00000000
#> 2 2 0.91564742 0.47827629
#> 3 3 0.03897943 0.02314612
#>
#> attr(,"class")
#> [1] "tree_dat" "list"Now it is straightforward to map over each of these trees to obtain the total expected values
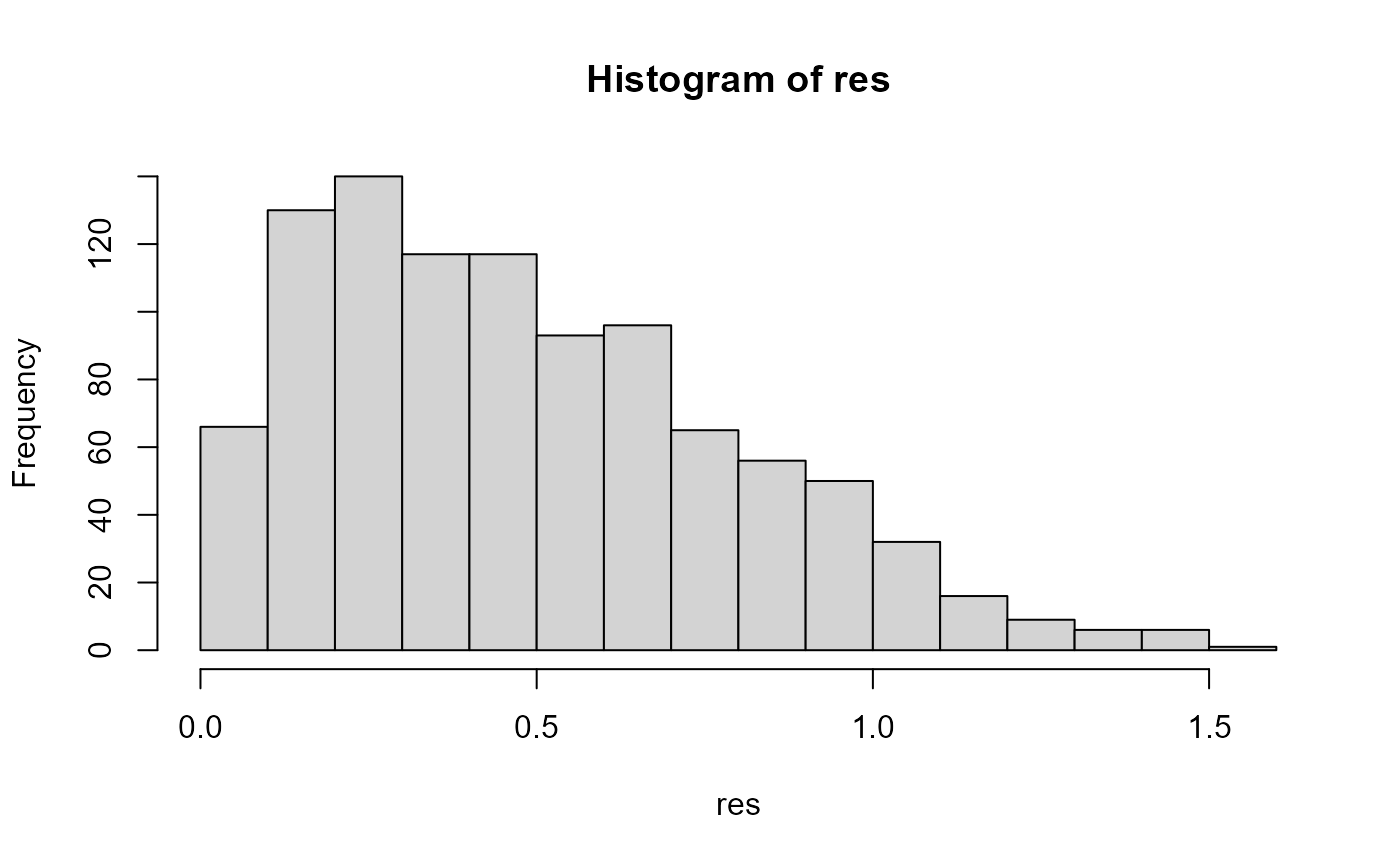
res <- map_dbl(tree_dat_sa, dectree_expected_values)
head(res)
#> [1] 0.4395898 0.4388347 0.9441533 0.6099253 0.5126502 0.4441804
hist(res, breaks = 20)